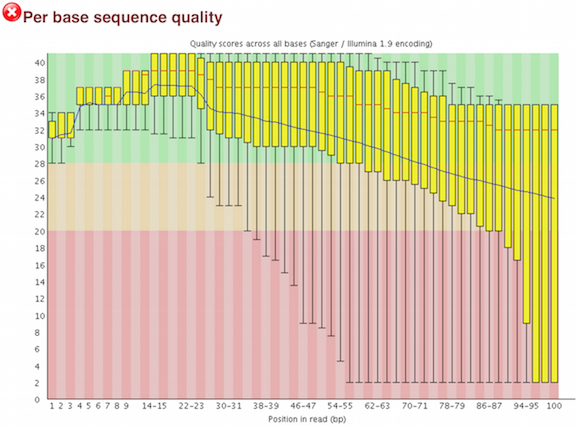
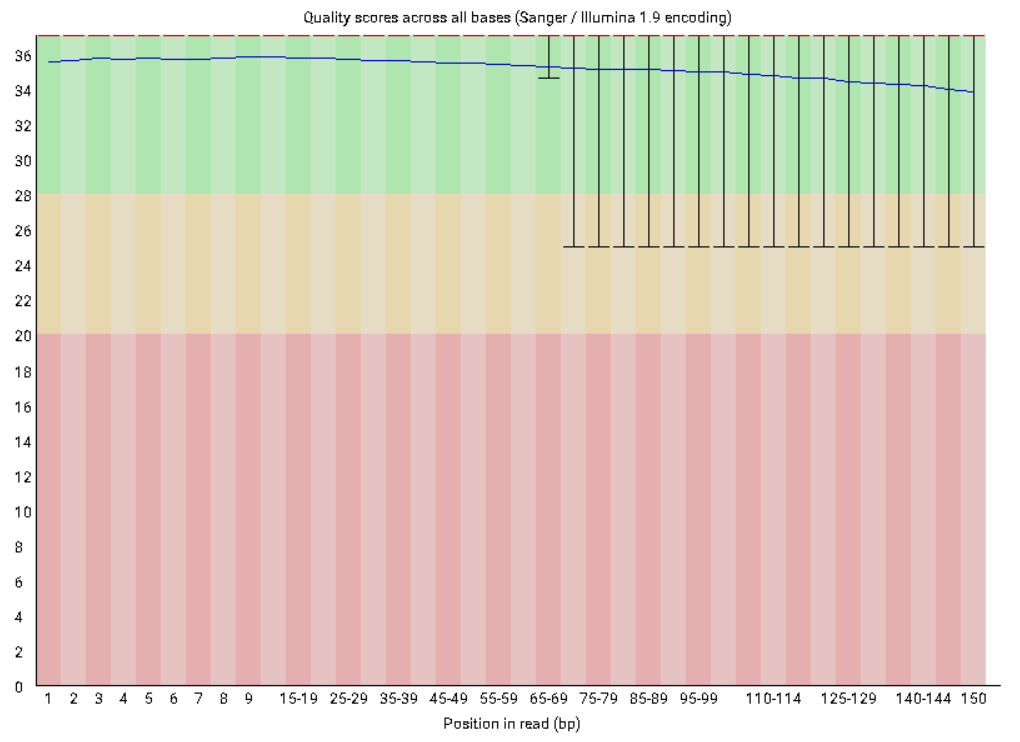
A representative example of per base sequence quality for RNA-Seq data... | Download Scientific Diagram
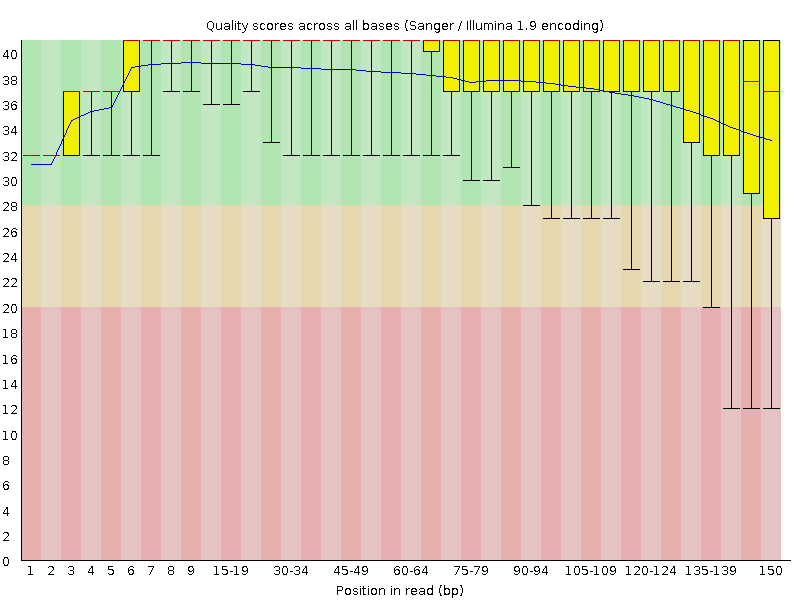
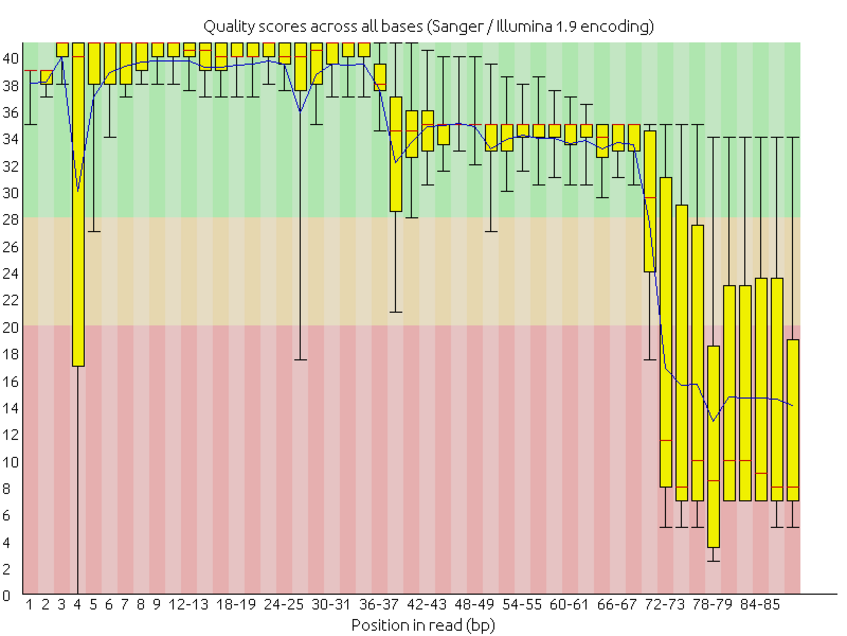
![Introduction to Next Generation Sequencing Bioinformatics”] | [“A Tufts University Research Technology Workshop”] Introduction to Next Generation Sequencing Bioinformatics”] | [“A Tufts University Research Technology Workshop”]](https://rbatorsky.github.io/intro-to-ngs-bioinformatics/img/fastqc_before_trim.png)
Introduction to Next Generation Sequencing Bioinformatics”] | [“A Tufts University Research Technology Workshop”]

Per base sequence quality from Illumina dataset.On the left bad quality... | Download Scientific Diagram

Quality control measures. (a) Per base sequence quality whisker plot:... | Download Scientific Diagram
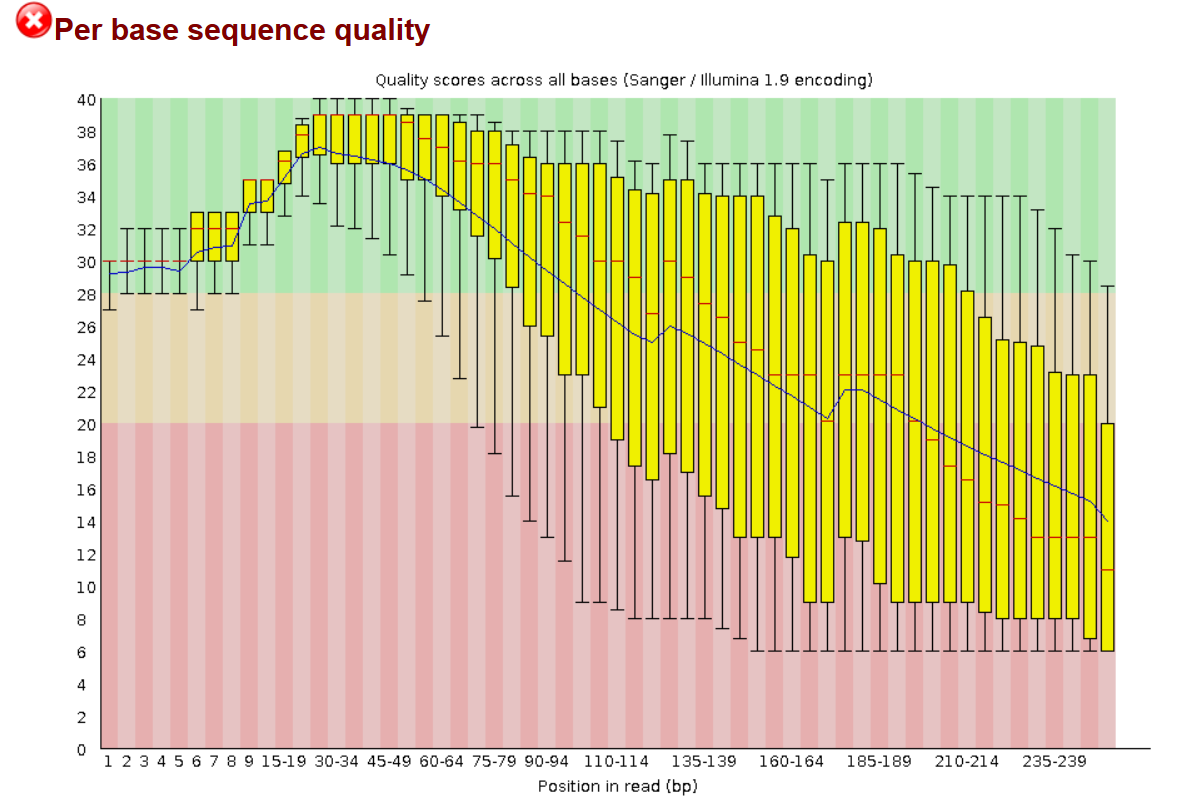
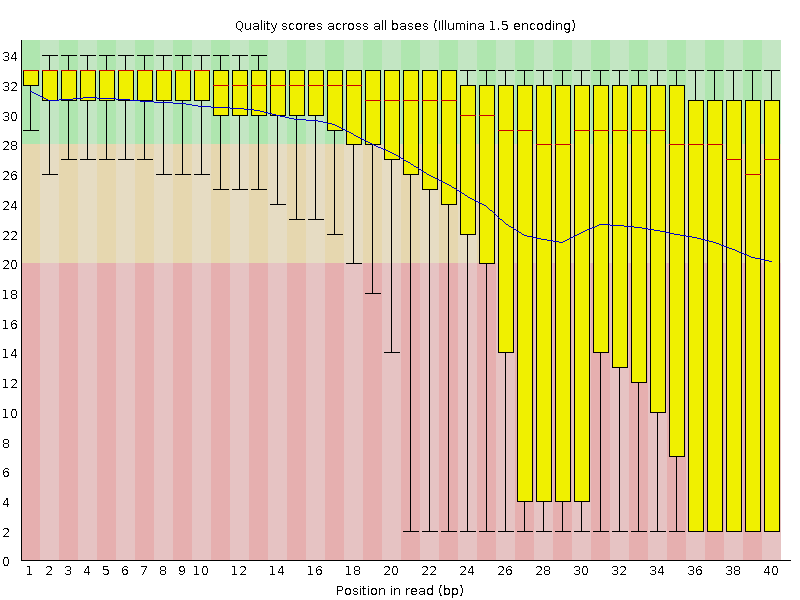
Analysing raw sequencing reads with FASTQC for quality control and filtering | by shilparaopradeep | Medium

SciBear🇺🇦 on Twitter: "(1/7) A quality control tool for raw #sequence data. Using #FastQC you may check: 🚀 Per base sequence #quality (do you see a drop in sequencing quality near the
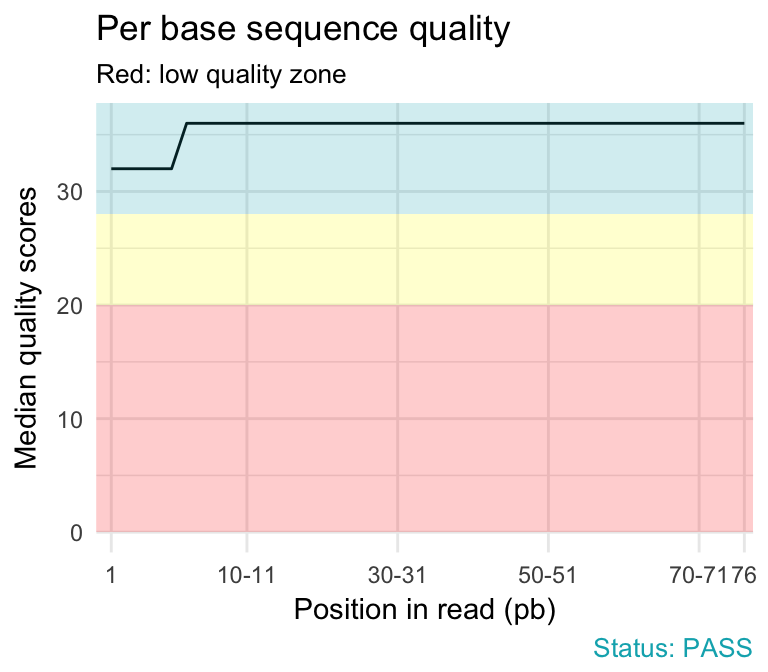
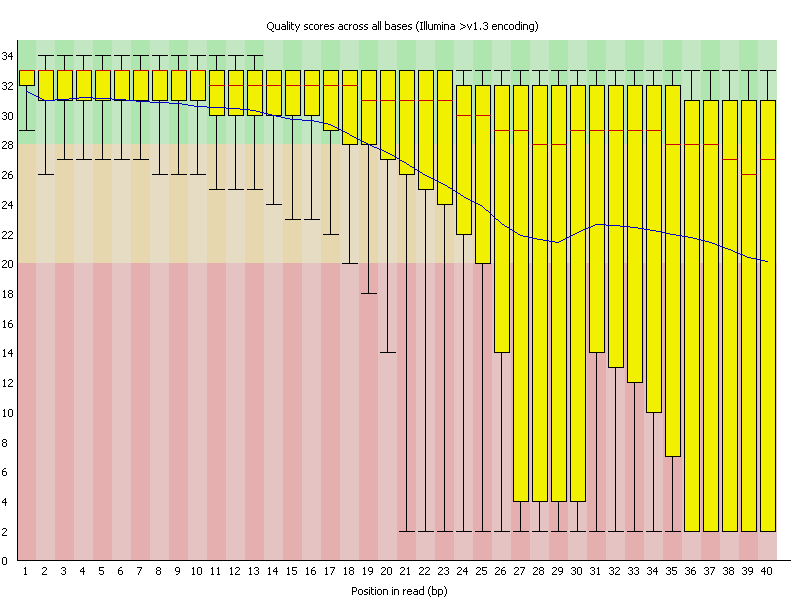
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples - Easy Guides - Wiki - STHDA
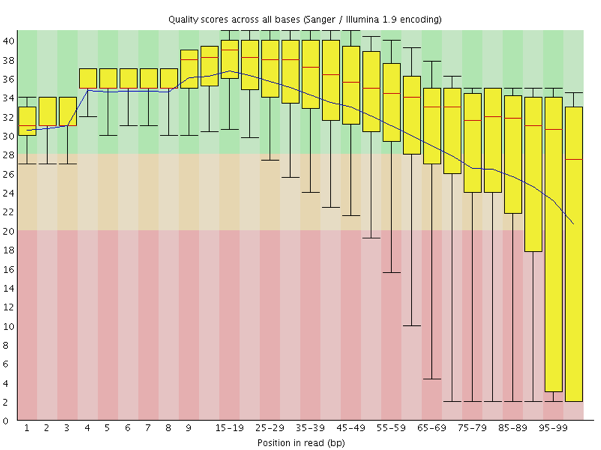
FASTQ Quality Assurance Tools - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis